Urine Exosome RNA Isolation Kit
A rapid procedure for the isolation of exosomal RNA from urine samples
For research use only and NOT intended for in vitro diagnostics.
A rapid procedure for the isolation of exosomal RNA from urine samples




For research use only and NOT intended for in vitro diagnostics.
A rapid procedure for the isolation of exosomal RNA from urine samples
Register today to receive an exclusive 15% off* on your first order.
This kit provides a rapid spin column procedure for the isolation of exosomal RNA from urine samples. Users can simultaneously concentrate and isolate high quality exosomal RNA, including microRNA, for use in sensitive downstream assays such as RT-PCR, qRT-PCR, NGS, microarrays and more. The protocol can be completed in under 50 minutes. Urine volumes of 1 to 10 mL can be processed easily and rapidly. All sizes of RNA are recovered at an equal rate without the need for using hazardous chemicals like phenol.
Background
Exosomes are 40 - 150 nm membrane vesicles, which are secreted by most cell types. Exosomes can be found in saliva, blood, urine, amniotic fluid and malignant ascite fluids, among other biological fluids. These vesicles act as cellular messengers, conveying information to distant cells and tissues within the body. The exosomes contain cell-specific proteins, lipids and RNAs, which are transported to other cells, where they can alter function and/or physiology. These exosomes may play a functional role in mediating adaptive immune responses to infectious agents and tumours, tissue repair, neural communication and transfer of pathogenic proteins. Recent work has demonstrated the presence of distinct subsets of microRNAs within exosomes which can inform about the cell type from which the exosomes are secreted. For this reason, exosomal RNAs may serve as biomarkers for various diseases including cancer. As the RNA molecules encapsulated within exosomes are protected from degradation by RNases they can be efficiently recovered from biological fluids, such as urine.
Figure 1. Detection of Urine Exosomal RNA Isolated from Different Urine Volumes. Norgen's Urine Exosome RNA Isolation Kit was used to isolate urine exosomal RNA from different urine volumes ranging from 1 to 15 mL. The purified urine exosomal RNA was then used as the template in an RT-qPCR reaction to detect the human 5S rRNA. 3 μL of the isolated urine exosomal RNA was used as the template in the RT step, and 3 µL from the RT step was used in the qPCR reaction. As it can be seen, the qPCR was able to successfully detect and amplify the 5S rRNA from exosomal RNA isolated from different urine volumes, indicating its high quality.
Figure 2. Effective and Consistent Detection of Urine Exosome RNA. Norgen's Urine Exosome RNA Isolation Kit can effectively isolate RNA from urine. Urine exosome RNA was isolated from 5 mL of human cell-free urine using Norgen's Urine Exosome RNA Isolation Kit (green), ExoQuick-TC Exosome Precipitation Reagent (blue) and Amicon® Ultra-15 Filter (blue). Stem loop RT-qPCR using primers specific to miR-21 and miR-16 as well as the housekeeping 5S rRNA was performed. In brief, three microliters of the 100 µL isolated RNA was then subjected to a 20 µL reverse transcription reaction using 5S rRNA, miR-21 and miR-16 stem-loop reverse primer or reverse primer. 3 μL from the reverse transcription reaction was used in a 20 µL real-time PCR reaction with primers to detect the human miR-21, human miR-16 and the 5S rRNA. Norgen's Urine Exosome RNA Isolation Kit is the only product that showed consistent detection of all tested transcripts with the highest quality.
Figure 3. High Diversity and Consistency of miRNA Recovered from Urine. Mid-stream urine was collected from three different healthy individuals. RNA was isolated from 20 mL of each sample using Norgen's Urine Exosome RNA Purification Kit (Cat. 47200). Small RNA Libraries were then generated using an Illumina TruSeq Small RNA Library Preparation Kit and subsequently sequenced on an Illumina MiSeq system. The normalized read counts of the mapped miRNAs were then compared between each individual sample. The scatter plots showed that the miRNA diversity was highly conserved among all individuals tested (R2 ≥ 0.92). This suggests that Norgen's RNA purification workflow recovers consistent profiles of small RNAs. This could enhance the possibility of detecting potential urine-based small RNA biomarkers.
Figure 4. High Degree of Overlapping of miRNA profile between Urine and Plasma. Plasma and mid-stream urine was collected from three different healthy individuals. RNA was isolated from 20 mL of each urine sample using Norgen's Urine Exosomal RNA Purification Kit (Cat. 47200) and 200 µL of each plasma sample using Norgen's Plasma/Serum RNA Purification Mini Kit (Cat. 55000). Small RNA Libraries were then generated using an Illumina TruSeq Small RNA Library Preparation Kit and subsequently sequenced on an Illumina MiSeq system. The list of mapped miRNAs from plasma and urine from the same individual was then compared. The Venn diagrams showed that the miRNA profiles have a high degree of overlapping between urine and plasma within each individual.
Figure 5. Increased Diversity of Other Small RNA Species (Piwi-Interacting) in Urine. Plasma and mid-stream urine was collected from three different healthy individuals. RNA was isolated from 20 mL of each urine sample using Norgen's Urine Exosomal RNA Purification Kit (Cat. 47200) and 200 µL of each plasma sample using Norgen's Plasma/Serum RNA Purification Mini Kit (Cat. 55000). Small RNA Libraries were then generated using an Illumina TruSeq Small RNA Library Preparation Kit and subsequently sequenced on an Illumina MiSeq system. The above chart shows that the relative proportion of piwi-interacting RNA (piRNA) was consistently higher in urine.
Figure 6. Western blot analysis of Norgen-enriched uEVs (urinary Extracellular Vesicles) from BPH (benign prostatic hyperplasia) and PCa (Pancreatic Cancer) urine samples. (Royo F, et al. Different EV enrichment methods suitable for clinical settings yield different subpopulations of urinary extracellular vesicles from human samples. Journal of Extracellular Vesicles. 2016;5:10.)
Figure 7. EV pellets recovered by Norgen’s Urine Exosome RNA Isolation Kit (Cat. 47200) kit were visualised by EM (Electron Microscopy) for sizing (Crossland RE, Norden J, Bibby LA, Davis J, Dickinson AM. Evaluation of optimal extracellular vesicle small RNA isolation and qRT-PCR normalisation for serum and urine. J Immunol Methods. 2016 Feb;429:39-49.)
|
Kit Specifications
|
|
| Minimum Urine Input |
1 mL
|
| Maximum Urine Input |
10 mL
|
| Size of RNA Purified |
Small exosomal RNA species
|
| Time to Complete Purification |
~ 50 minutes
|
Storage Conditions and Product Stability
All buffers should be kept tightly sealed and stored at room temperature. This kit is stable for 2 years after the date of shipment.
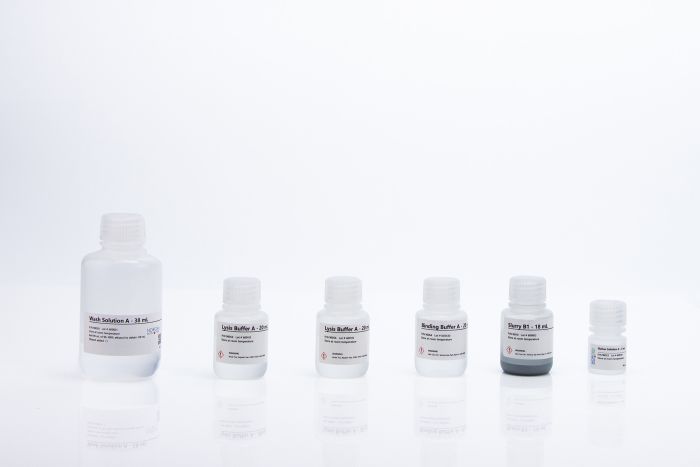
| Component | Cat. 47200 (50 preps) |
|---|---|
| Slurry B1 | 18 mL |
| Binding Buffer A | 20 mL |
| Lysis Buffer A | 2 x 20 mL |
| Wash Solution A | 38 mL |
| Elution Solution A | 6 mL |
| Mini Filter Spin Columns | 50 |
| Collection Tubes | 50 |
| Elution Tubes (1.7 mL) | 50 |
| Product Insert | 1 |
| Title | Maternal-Infant Factors in Relation to Extracellular Vesicle and Particle miRNA in Prenatal Plasma and in Postpartum Human Milk |
| Citation | International Journal of Molecular Sciences. 2024. |
| Authors | by Meghan E. Muse 1,*ORCID,David A. Armstrong 2,3ORCID,Anne G. Hoen 1,4,Diane Gilbert-Diamond 1ORCID,Jiang Gui 4,Thomas J. Palys 1,Frederick W. Kolling 5,Brock C. Christensen 1,6,Margaret R. Karagas 1 and Caitlin G. Howe |
| Title | Development of a robust and generalizable algorithm "gQuant" for accurate normalizer gene selection in qRT-PCR analysis |
| Citation | Scientific Reports 2024. |
| Authors | Abhay Kumar Pathak, Sukhad Kural, Shweta Singh, Lalit Kumar, Mahima Yadav, Manjari Gupta, Parimal Das & Garima Jain |
| Title | gQuant: A Robust and Generalizable Algorithm for Identifying Normalizer Genes in qRT-PCR Data: A Case Study on Urinary Exosomal miRNAs |
| Citation | bioRxiv 2023. |
| Authors | Abhay Kumar Pathak, Sukhad Kural, Shweta Singh, Lalit Kumar, Mahima Yadav, Manjari Gupta, Parimal Das, Garima Jain |
| Title | Proximal tubule-derived exosomes contribute to mesangial cell injury in diabetic nephropathy via miR-92a-1-5p transfer |
| Citation | Cell communication and signaling : CCS 2023. |
| Authors | Yi-Chun Tsai, Mei-Chuan Kuo, Wei-Wen Hung, Ping-Hsun Wu, Wei-An Chang, Ling-Yu Wu, Su-Chu Lee & Ya-Ling Hsu |
| Title | Urinary exosomal miRNA signature of IgA nephropathy: a case-control study |
| Citation | scientific reports 2023. |
| Authors | Mythri Shankar, Aditya Shetty, Madhura N.S., Sreedhara C.G., Kishan A. & Karthik Tennankore |
| Title | Urinary exosome-derived microRNAs reflecting the changes of renal function and histopathology in dogs |
| Citation | Scientific Reports 2017. |
| Authors | Ichii, O., Ohta, H., Horino, T., Nakamura, T., Hosotani, M., Mizoguchi, T., ... & Sasaki, N |
| Title | Urinary extracellular vesicles for RNA extraction: optimization of a protocol devoid of prokaryote contamination |
| Citation | Journal of Extracellular Vesicles 2016. |
| Authors | Tataruch-Weinert, D., Musante, L., Kretz, O., & Holthofer, H |
| Title | Vesicle-MaNiA: extracellular vesicles in liquid biopsy and cancer |
| Citation | ScienceDirect 2016. |
| Authors | Torrano, V., Royo, F., Peinado, H., Loizaga-Iriarte, A., Unda, M., Falcón-Perez, J. M., & Carracedo, A |
| Title | Investigation on torquetenovirus (TTV) microRNA transcriptome in vivo |
| Citation | Virus Research 2016. |
| Authors | Vignolini, T., Macera, L., Antonelli, G., Pistello, M., Maggi, F., & Giannecchini, S |
| Title | Urinary exosomal microRNA panel unravels novel biomarkers for diagnosis of type 2 Diabetic Kidney Disease |
| Citation | Journal of Diabetes and its Complications 2016. |
| Authors | Eissa, S., Matboli, M., Aboushahba, R., Bekhet, M. M., & Soliman, Y |