Next-Generation Sequencing Approach to Investigate Urinary microRNA in Prostate Cancer
Urinary miRNA Biomarkers
For the past several years, there has been evolving evidence that alteration in gene expression of microRNAs (miRNAs) is associated with several diseases, including human malignancies such as prostate cancer (PCa). miRNAs are small non-coding RNA molecules (19-25 nt long) that target messenger RNAs (mRNAs) to repress protein expression.1 They regulate various biological processes such as the cell cycle, cell proliferation, apoptosis, migration, invasion, and metastasis.2 Alteration in this biological process including miRNA dysregulation could provide a crucial indication during the early stages and progression of cancer diseases. In 2018, Guelfi and colleagues investigated the use of urinary miRNA as a non-invasive biomarker for the diagnosis of PCa through NGS analysis. They were the first scientists to conduct a pilot study using Illumina Deep Sequencing to evaluate the entire miRNA profile of urine exfoliated prostate cells (UEPCs) in PCa patients.3
Prostate Cancer Detection
Currently, a prostate biopsy is the gold standard procedure for prostate cancer diagnosis after a suspicious indication of the disease through a serum Prostate-Specific Antigen (PSA) test and the Digital Rectal Examination (DRE).3,4 PSA is used as the primary trademark for prostate cancer screening. However, this antigen-based test has many limitations due to the non-cancer specificity results; elevated levels of PSA can be indicative of several other diseases, leading to an over-preliminary diagnosis (false-positive result) of PCa and an increased number of needless biopsies.5 DRE, on the other hand, is a physical examination used mostly as a second examination for further investigation of the malignancy, but due to the low reproducibility and sensitivity, is used together with PSA for prostate cancer detection.6
Following an abnormal PSA and DRE test, a prostate biopsy is done to diagnose the presence of PCa with a higher degree of confidence. One of the major problems of prostate biopsy, aside from being a highly invasive tool for cancer detection, is the risk of false-negative which can result in patients having to go through the procedure again.3 In addition, patients that undergo the biopsy procedure can experience other complications, such as infection, which is the common reason for hospitalization after the biopsy.4
Studies to identify novel PCa biomarkers as a strategy to overcome the limitation of the PSA test have been the research focus for many scientists, but so far many of these test results can not be reproduced, and the protocols are too complicated for use in a clinical setting.7 Consequently, it is important to identify and validate novel PCa biomarkers that can be tested clinically and with precise reproducibility.8
It has been suggested that miRNA analysis in biofluids such as urine is a great method for clinical application. Urine is a non-invasive sample that can be used to find biomarkers that represent the genomic spectrum of the entire tumor, instead of only a local tumor sample.9 Studies that investigate urinary cell-free miRNA encourage the use of urine-based liquid biopsy samples as a favorable approach to diagnosing prostate cancer, and the rapid process in miRNAs profiling technology provides a great tool to study the expression level of miRNA through NGS.3,10
NORBLOG
Want to hear more from Norgen?
Join over 10,000 scientists, bioinformaticians, and researchers who receive our exclusive deals, industry updates, and more, directly to their inbox.
For a limited time, subscribe and SAVE 10% on your next purchase!
SIGN UP
Urinary miRNA Isolation for PCa Detection
To examine the miRNA expression level of urine exfoliated prostate cells in prostate cancer patients, Guelfi et al. selected male patients that were clinically diagnosed with PCa for the NGS study, and the results were validated by comparing it with healthy control subjects through qPCR analysis. The researchers collected the first void of urine using Norgen’s Urine Preservative to preserve nucleic acids and proteins at room temperature, and the total RNA was extracted using Norgen’s Urine Exfoliated Cell and Bacteria RNA Purification Kit.
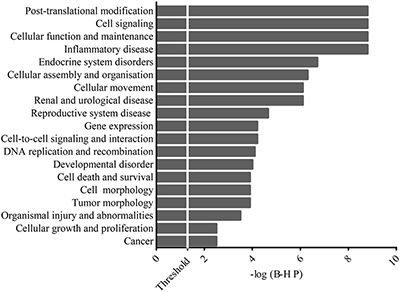
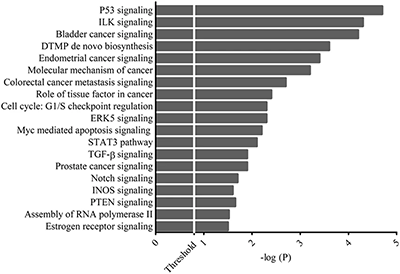
Once the RNA was purified, the miRNA profile of the PCa samples was examined using Illumina small RNA seq, and the top 62 expressed miRNAs target genes were selected for the gene ontology (GO) and pathway analyses to investigate the relationship between PCa and the selected genes. The functional analysis showed that most of the top 62 miRNAs were involved in diseases and other biological processes (Figure 1A)3, and the core function-specific analysis showed that some genes were involved in prostate cancer signaling (Figure 1B).3
After the predictive activities of selected PCa miRNAs by NGS, Guelfi et al. validated the results by comparing the expression level of the PCa miRNAs with the healthy control group using the miRNA ever identified in humans (lethal-7 or let 7), through qPCR (Figure 3).3
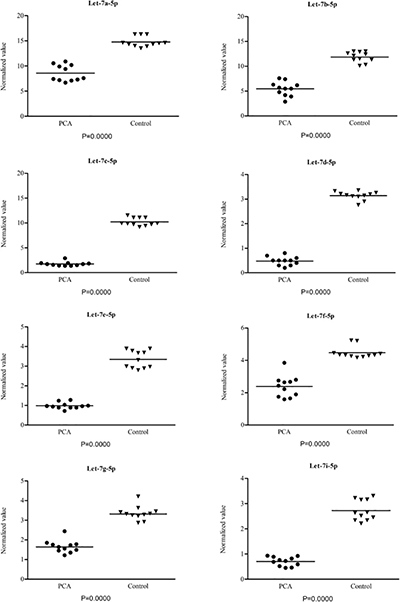
In this study, Guelfi et al. provided supporting evidence of the value of miRNA for prostate cancer detection, and the use of NGS Illumina Deep Sequencing to detect and identify novel miRNAs. Guelfi et al. concluded that the profile of the let-7 family gene in the urine exfoliated prostate cells could be an easy and affordable non-invasive tool for the detection and management of prostate cancer.
-
Massillo, C., Dalton, G. N., Farré, P. L., De Luca, P., & De Siervi, A. (2017). Implications of microRNA dysregulation in the development of prostate cancer. Reproduction, 154(4), R81-R97.
-
Ruan, K., Fang, X., & Ouyang, G. (2009). MicroRNAs: novel regulators in the hallmarks of human cancer. Cancer letters, 285(2), 116-126.
-
Guelfi, G., Cochetti, G., Stefanetti, V., Zampini, D., Diverio, S., Boni, A., & Mearini, E. (2018). Next generation sequencing of urine exfoliated cells: an approach of prostate cancer microRNAs research. Scientific reports, 8(1), 1-8.
-
Loeb, S., Vellekoop, A., Ahmed, H. U., Catto, J., Emberton, M., Nam, R., ... & Lotan, Y. (2013). Systematic review of complications of prostate biopsy. European urology, 64(6), 876-892.
-
Hafron, J. M., Yu, H., Juang, A., Vuong, D., Kamer, S., Carbonell, L., ... & Rajan, R. (2017). New developments in prostate cancer screening using a novel cancer-specific, non-PSA biomarker assay derived from autoantibody signatures. J Med Ther.
-
Partin, A. W., Wein, A. J., Kavoussi, L. R., Peters, C. A., & Dmochowski, R. R. (2020). Campbell Walsh Wein Urology, E-Book. Elsevier Health Sciences.
-
Liu, D. F., Wu, J. T., Wang, J. M., Liu, Q. Z., Gao, Z. L., & Liu, Y. X. (2012). MicroRNA expression profile analysis reveals diagnostic biomarker for human prostate cancer. Asian Pacific Journal of Cancer Prevention, 13(7), 3313-3317.
-
Byun, Y. J., Piao, X. M., Jeong, P., Kang, H. W., Seo, S. P., Moon, S. K., ... & Kim, W. J. (2021). Urinary microRNA-1913 to microRNA-3659 expression ratio as a non-invasive diagnostic biomarker for prostate cancer. Investigative and clinical urology, 62(3), 340.
-
Endzeliņš, E., Melne, V., Kalniņa, Z., Lietuvietis, V., Riekstiņa, U., Llorente, A., & Linē, A. (2016). Diagnostic, prognostic and predictive value of cell-free miRNAs in prostate cancer: a systematic review. Molecular cancer, 15(1), 1-13.
-
Mlcochova, H., Hezova, R., Stanik, M., & Slaby, O. (2014, January). Urine microRNAs as potential noninvasive biomarkers in urologic cancers. In Urologic Oncology: Seminars and Original Investigations (Vol. 32, No. 1, pp. 41-e1). Elsevier.