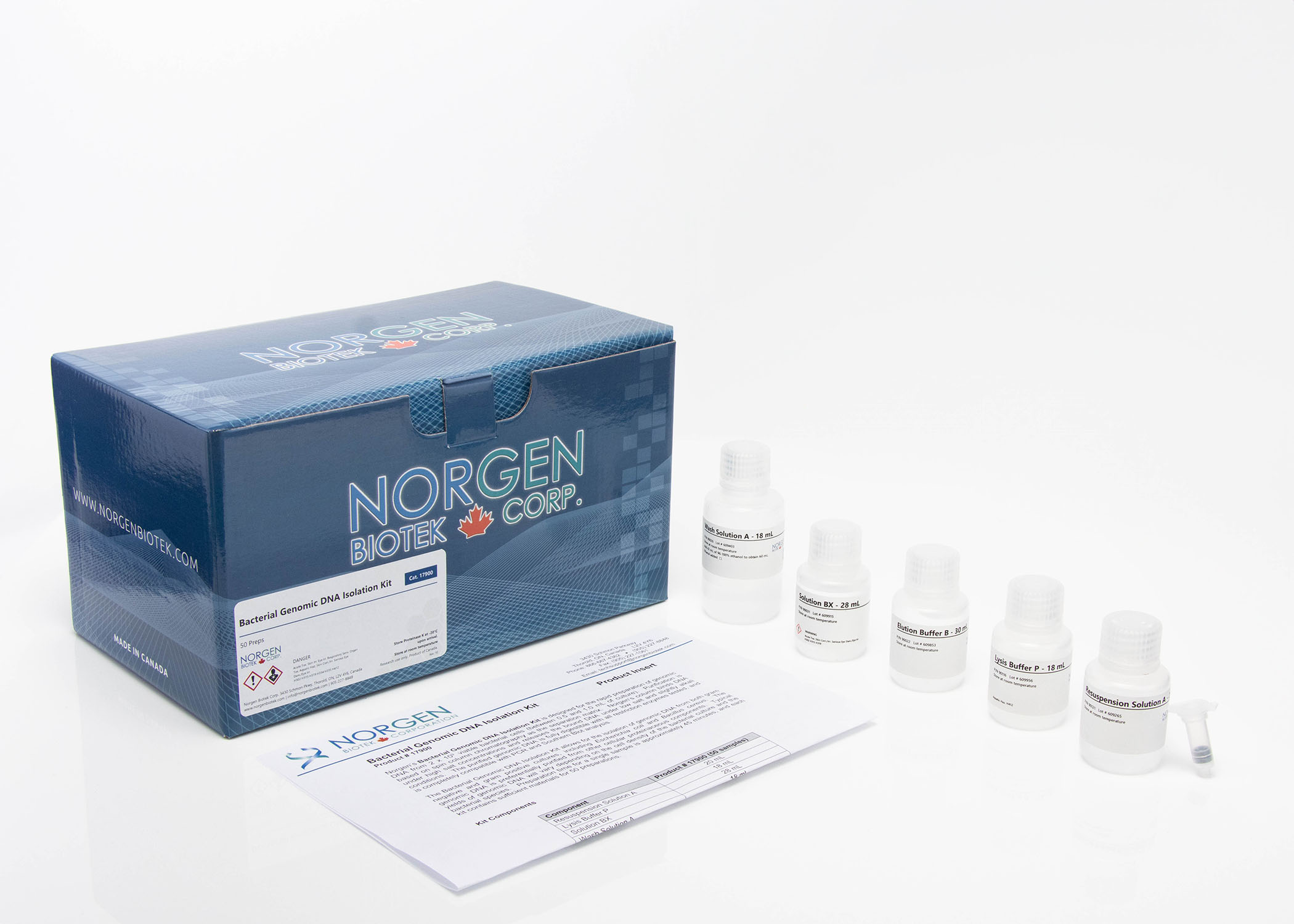
Phage DNA Isolation Kit
For the rapid purification of total DNA from bacteriophages.



For research use only and NOT intended for in vitro diagnostics.
Phage DNA Isolation Kit
For the rapid purification of total DNA from bacteriophages.
Register today to receive an exclusive 15% off* on your first order.
Features and Benefits
- Isolate high quality DNA from a broad variety of phage strains
- High yields of total DNA
- Fast and easy processing using a rapid spin-column format
- No phenol or chloroform extractions or cesium chloride banding required
- High yields of DNA recovered 3-15 µg DNA from 108-1010 pfu/ mL of enriched phages
This kit provides a rapid spin column method for the purification of total DNA from a broad spectrum of bacteriophages propagated in bacteria grown in liquid cultures. The DNA is isolated without the use of phenol, chloroform or cesium chloride banding procedures. The spin-column based procedure is rapid and can be completed in less than 45 minutes. The kit is highly efficient for processing small volumes of phage supernatant (500 µL – 1 mL) and with the optional DNase and Proteinase K treatments phage DNA yields are maximized while host DNA contamination is minimized. Purified total phage DNA is of the highest integrity, and can be used in a number of downstream applications including PCR, qPCR, Restriction Fragment Length Polymorphism (RFLP), sequencing, cloning, Southern Blot and more.
Details
Supporting Data
Figure 1. Effective Host Genomic DNA Removal without Reducing Phage DNA Yield. Total DNA was isolated from four enriched phage cultures using Norgen's Phage DNA Isolation Kit. A DNase I pre-treatment was performed prior to adding the provided Lysis Buffer. Briefly, 20 units of DNase I was added to 1 mL of enriched phage culture and the mixture was incubated at room temperature for 20 minutes. After the DNAase I treatment the procedure was followed. As a control, DNA was isolated from aliquots of the same 4 cultures using Norgen’s Phage DNA Isolation Kit without performing the DNase I treatment. For DNA analysis 10 µL of each 50 µL elution was loaded onto a 1X TAE agarose gel. As it can be seen, the phage DNA was safely protected from the DNase I treatment by its coat protein, while the host genomic DNA was efficiently degraded by the DNase I. Thus the DNase I pre-treatment resulted in less host gDNA contamination in the final phage elution without influencing the total phage DNA yield. Lane M is Norgen's Highranger 1 kb DNA Ladder (Cat. 11900)
Figure 2. Optional Proteinase K Treatment Improves DNA Yield for Certain Phage Strains. Total DNA was isolated with and without the optional Proteinase K treatment using Norgen's Phage DNA Isolation Kit. Briefly, 4 µL of Proteinase K (20 mg/mL) was added to 1 mL of enriched phage culture and incubated at 55°C for 15 minutes with the phage Lysis Buffer. After the Proteinase K treatment the procedure was followed. As a control, DNA was isolated from aliquots of the same 8 cultures using Norgen's Phage DNA Isolation Kit without performing the Proteinase K treatment. For DNA analysis 10 µL of each 50 µL elution was loaded onto a 1X TAE agarose gel and the yield of DNA was compared from the eight different phage types (lane 1 to 8). As it can be seen, the optional treatment of Proteinase K improved the phage DNA yield in Lanes 2, 5 and 6 dramatically. Lane M is Norgen's Highranger 1 kb DNA Ladder (Cat. 11900)
|
Kit Specifications
|
|
|
Column Binding Capacity
|
50 µg
|
|
Maximum Column Loading Volume
|
650 µL
|
|
Size of DNA Purified
|
All sizes
|
| Maximum Amount of Starting Material |
1 x 1010 pfu/mL enriched phages
|
| Average Yield* |
3-15 µg DNA from 108-1010 pfu/mL
of enriched phages |
| Time to Complete 10 Purifications |
45 minutes
|
* Average yields will vary depending upon a number conditions used and developmental stage.
Storage Conditions and Product Stability
All solutions should be kept tightly sealed and stored at room temperature. This kit is stable for 1 year after the date of shipment.
| Component | Cat. 46800 (50 preps) | Cat. 46850 (100 preps) |
|---|---|---|
| Lysis Buffer B | 40 mL | 2 x 40 mL |
| Wash Solution A | 38 mL | 2 x 38 mL |
| Elution Buffer B | 8 mL | 2 x 8 mL |
| Spin Columns | 50 | 100 |
| Collection Tubes | 50 | 100 |
| Elution Tubes (1.7 mL) | 50 | 100 |
| Product Insert | 1 | 1 |
Documentation
FAQs
Spin Column
Column clogging may occur due to one or more of the following reasons:
- Centrifugation speed was too low or spin time was inadequate.
Check the centrifuge to ensure that it is capable of generating the required RPMs. Sufficient centrifugal force is required to move the liquid through the resin. Also, ensure that the correct spin times are followed. Spin for an additional minute if necessary.
- Bacterial debris in the lysate.
Ensure that the starting material is clarified phage supernatant. Remove bacterial debris from the initial phage supernatant by centrifugation at 10,000 × g for 5 minutes before beginning the protocol.
- The lysate/binding solution mixture is not homogeneous.
To ensure a homogeneous solution, vortex for 10-15 seconds before applying the lysate to the spin column.
- Centrifuge temperature is too low.
Ensure that the centrifuge remains at room temperature throughout the procedure. Temperatures below 20℃ may cause precipitates to form that can cause the columns to clog.
The yield of genomic DNA may be lower than expected due to the following:
- Ineffective propagation of phage and initial lysis step.
Refer to the manufacturer's recommendations for the propagation of the phage, including proper titer for inoculation, growth conditions, and bacterial host.
- Incomplete lysis of cells.
Ensure that the incubation was performed for 15 minutes at 65℃ after the addition of Lysis Buffer B. Also, perform optional digestion with Proteinase K in Step 1b during Lysate Preparation.
- Ethanol was not added to the Wash Solution A.
Ensure that 90 mL of 96 - 100% ethanol is added to the supplied Wash Solution A prior to use.
- The DNA elution is incomplete.
Ensure that all the DNA is eluted. If elution buffer remains in the column, use 14,000 g for the second centrifuge.
If the DNA does not perform well in downstream applications, it may be due to one or more of the following:
- DNA was not washed three times with the provided Wash Solution A.
Traces of salt from the binding step may remain in the sample if the column is not washed three times with the Wash Solution A. Salt may interfere with downstream applications and thus must be washed from the column.
- Ethanol carryover.
Ensure that the dry spin under the Column Wash procedure is performed in order to remove traces of ethanol prior to elution. Ethanol is known to interfere with many downstream applications.
Host genomic DNA can contaminate the phage DNA. In order to eliminate host genomic DNA contamination in the phage DNA elution, it is recommended that a DNase I treatment is performed at the beginning of the lysate preparation procedure (see Section 1, Optional DNase Treatment).
Yes, the kit can isolate both ssDNA and dsDNA phage genomes.
Citations
| Title | Discovery and characterisation of new phage targeting uropathogenic Escherichia coli |
| Citation | Prepriint BIORXIV 2024. |
| Authors | Shahla Asgharzadeh Kangachar1 , Dominic Y. Logel2,3, Ellina Trofimova2,3, Hannah X Zhu2,3, Julian Zaugg1 , Mark A. Schembri4,5, Karen D. Weynberg1 , Paul R. Jaschke2,3* |
| Title | In vitro evaluation of two novel Escherichia bacteriophages against multiple drug resistant avian pathogenic Escherichia coli |
| Citation | BMC Infectious Diseases 2024. |
| Authors | Mobina Karami, Ali Goudarztalejerdi, Abdolmajid Mohammadzadeh & Enayat Berizi |
| Title | Genomic and Proteomic Analysis of Six Vi01-like Phages Reveals Wide Host Range and Multiple Tail Spike Proteins |
| Citation | Viruses 2024. |
| Authors | Evan B. Harris , Kenneth K. K. Ewool , Lucy C. Bowden, Jonatan Fierro, Daniel Johnson, McKay Meinzer, Sadie Tayler and Julianne H. Grose |
| Title | Genomic analysis of a novel phage vB_SenS_ST1UNAM with lytic activity against Salmonella enterica serotypes |
| Citation | Diagnostic Microbiology and Infectious Disease 2024. |
| Authors | Gerardo E Rodea M b c, Edgar González-Villalobos h, María del Rosario Espinoza-Mellado d, Ulises Hernández-Chiñas a b, Carlos Alberto Eslava-Campos a b, José Luis Balcázar e f, José Molina-López |
| Title | Genome sequences of actinobacteriophages JorRay, Blocker23, Nibbles, and OlgasClover |
| Citation | Microbiology Resource Announcements 2024. |
| Authors | Breanna M. Baumgartner, Kayla A. Bono, Dawn R. McIntosh, Anna M. Vu, Chanel F. Adams, Brooklyn C. Benik, Jessica Chavez, Samantha J. Gresky, Andres Sotelo, Jordan I. Ray, Alexandra Peister, Kendra W. Kimberley, Chelsey C. McKenna, James R. Theoret, Earl J. Yoon https://orcid.org/0000-0002-7302-8819 earl.yoon@csn.edu, Erin J. Windsor |
| Title | A gradient phage-assisted continuous evolution method for screening suppressor tRNAs in Escherichia coli |
| Citation | New Biotechnology 2024. |
| Authors | Fan Wang a b 1, Li-Hua Liu b c 1, Zhenyu Wang a 1, Ao Jiang b 1, Yi-Rui Wu |
| Title | Complete genome sequence of Pseudomonas aeruginosa phage Knedl |
| Citation | Microbiology Resource Announcements 2024. |
| Authors | Enea Maffei,1,2 Christina Manner,1 Urs Jenal,1 Alexander Harms |
| Title | Characterization of two novel Salmonella phages having biocontrol potential against Salmonella spp. in gastrointestinal conditions |
| Citation | scientific reports 2024. |
| Authors | Characterization of two novel Salmonella phages having biocontrol potential against Salmonella spp. in gastrointestinal conditions |
| Title | Combinations of Bacteriophage Are Efficacious against Multidrug-Resistant Pseudomonas aeruginosa and Enhance Sensitivity to Carbapenem Antibiotics |
| Citation | Viruses 2024. |
| Authors | Christopher J. Kovacs 1,2,Erika M. Rapp 1,William R. Rankin 1,Sophia M. McKenzie 1,Brianna K. Brasko 1,Katherine E. Hebert 1,Beth A. Bachert 1ORCID,Andrew R. Kick 1ORCID,F. John Burpo 1ORCID andJason C. Barnhill |
| Title | Isolation and characterization of a novel phage P785, its combination with thermal processing for the biocontrol of Escherichia coli in milk |
| Citation | Food Safety and Health 2024. |
| Authors | Hongmei Yin | Jinli Wang | Likou Zou | Anjian Liang | Chenglin Zhu | Yu Fu | Juan Chen | Junni Tang |