A Possible Origin of Life
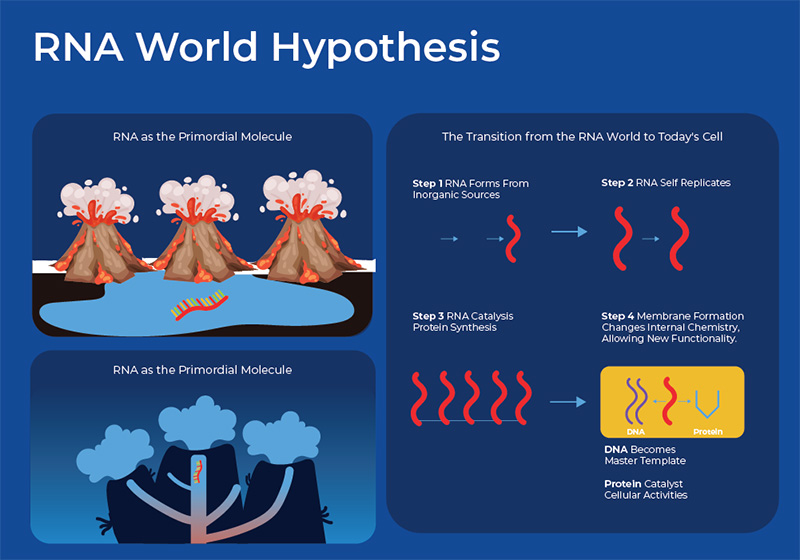
Philosophical, theological and scientific thoughts about the beginning of life have captivated societies for millenia. It is generally agreed upon that the earth is 4.5 billion years old, and had conditions conducive to life by about 4.3 billion years ago. Darwin is the scientific champion for the theory of evolution, and even the bible mentions some sequential process in which life on land was preceded by life in the water, and plant life preceded animal life. Regardless, it had to start somewhere. Now, the main scientific theory on the origin of life posits that it began with RNA forming in a primordial soup. In the “RNA world hypothesis”, RNA served a dual purpose of both encoding genetic information (that role has now been passed on to DNA), and performing catalytic functions (that role has now been passed on to protein enzymes).1
But how did the original RNA molecule arise? There are three main elements that are considered necessary to create RNA from the prebiotic chemical earth: an energy source, organic compounds, and water. Many different possible energy sources have been suggested, including: hydrothermal vents, electrical spark discharges, geothermal hot springs, ultraviolet radiation, and volcanic eruptions, alongside external sources such as meteoroids, asteroids, and comets. Some research suggests that these extra-terrestrial objects also brought chemical building blocks necessary for RNA synthesis, while others believe that all of the chemistry necessary for the formation of RNA was already present on earth. Various experiments that have replicated prebiotic chemical and physical parameters in Warm Little Ponds (WLP) or hydrothermal vents, have shown the creation of organic compounds from the chemical building blocks on earth. Along this continuum from the first RNA molecule to the first cell, initial steps include the formation of nucleotides, fatty acids, and various other biomolecules. Following this, encapsulated self-replicating information polymers emerged, paving the way for the development of cellular life. Although life may have started with a simple RNA molecule, new research is revealing that there now exists many different types of RNA that differ in their biogenesis, structure, function, and biological roles. This blog will explore the diversity of these RNA molecules.2
What is RNA
Ribonucleic acid (RNA) is a type of Nucleic Acids, one of the four main macromolecules necessary for all known forms of life.
RNA is a polymeric molecule composed of chains of nucleotides. Each nucleotide is composed of a ribose sugar group (DNA has deoxyribose instead) with an attached phosphate group, and one of four nitrogenous bases. Three out of the four nitrogenous bases are the same as those in DNA - Adenine, Cytosine, Guanine - however, instead of Thiamine (as in DNA), RNA has Uracil.
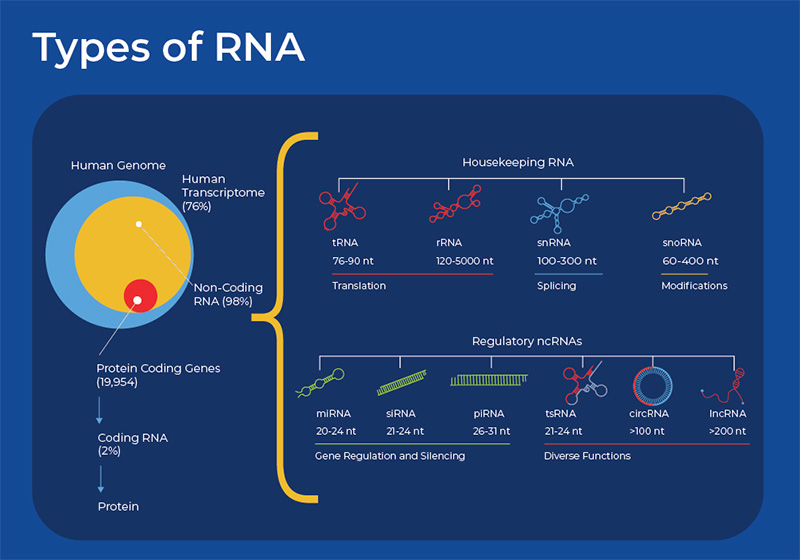
RNA is essential for most biological processes and its functions can be categorized into two main roles. Firstly, RNA can act as a messenger encoding the instructions from DNA and passing them to the ribosome for protein synthesis (messenger RNA). Secondly, the RNA itself can perform a function; these RNA are called non-coding RNA (ncRNAs). Sequencing of the human genome and transcriptome has revealed that about 76% of the genome is transcribed but only 2% actually codes for proteins. We are thus unsure about the function of a large proportion of the human genome, but it is also now clear that this 98% that is not protein-coding is not junk DNA, as was originally thought. The ncRNAs possess a wide variety of functions and are often involved in housekeeping functions. They perform important regulatory roles in many areas of development and disease.
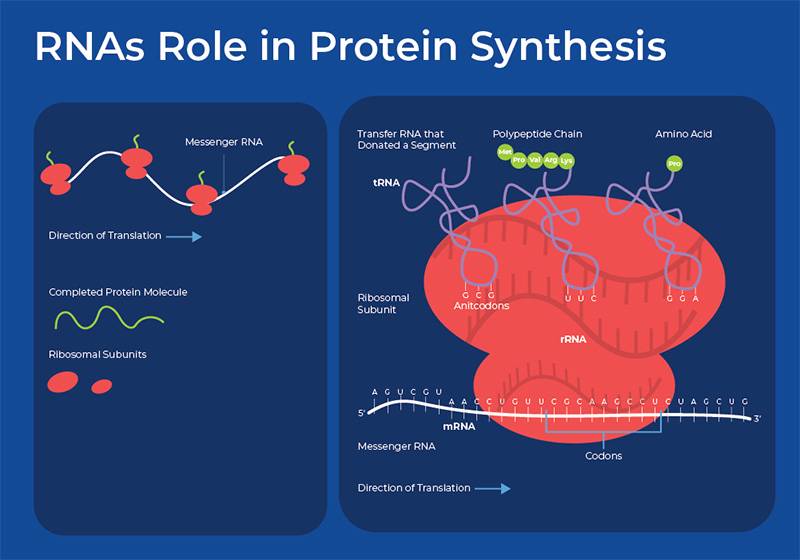
RNAs Role in Protein Synthesis
The main importance of RNA to the cell was originally thought to be in its role in making proteins. Proteins are the main workhorses of our cells, as they are involved in catalyzing reactions, transporting molecules, and providing structure to cells. There are three main types of RNA involved in protein synthesis: mRNA, tRNA and rRNA. mRNA is synthesized by the enzyme RNA polymerase using the sequence of bases in a strand of DNA to make a complementary strand of mRNA. The pre-mRNA is then processed - spliced, possibly edited, capped, and polyadenylated to increase its stability and increase the diversity of proteins it can code for. The mRNA then moves to the ribosome. The ribosome has two subunits, each composed of an rRNA molecule and its associated proteins. Once the mRNA enters the ribosome, another RNA species is involved - tRNA. tRNA molecules help deliver amino acids to the ribosome. Working together, the nucleotides in the mRNA dictate the order of the amino acids, the tRNA carries the amino acids, and the rRNA molecules that form the ribosome assemble the amino acids into the final protein.
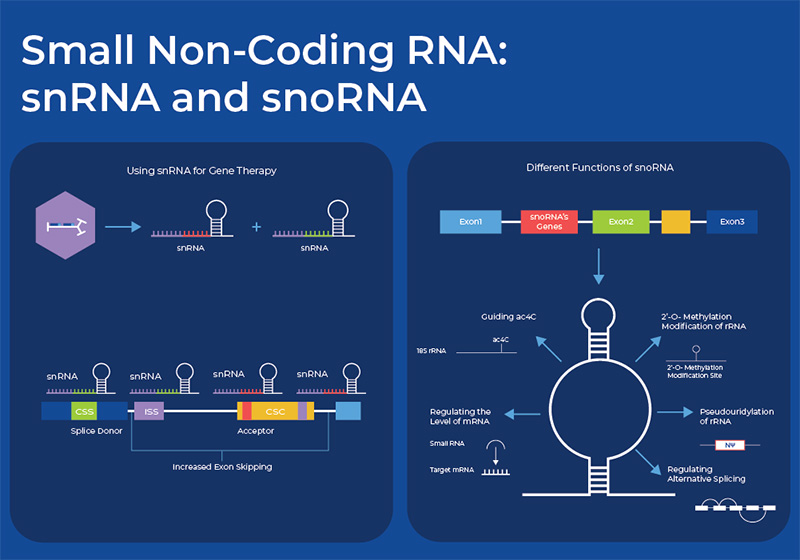
Small non-coding RNA: snRNA and snoRNA
This simplified view of the central dogma - DNA giving rise to protein through the function of mRNA, rRNA and tRNA - has become more complex in recent years as research into other small ncRNAs (sncRNA) has expanded. Upon closer inspection, protein synthesis and other housekeeping functions require a wider array of RNA species, including small nuclear RNA (snRNA) and small nucleolar (snoRNA).
Small nucleolar RNAs (snoRNAs)
Small nucleolar RNAs (snoRNAs) are small non-coding RNAs widely present in the nucleoli of eukaryotic cells, and have a length of 60–300 nt. They are encoded by intronic regions of both protein coding and non-protein coding genes. The main function of snoRNAs is in the complex process of chemical modifications that pre-RNAs undergo before they are mature rRNAs. snoRNAs have specific sequences that guide them to specific cellular RNAs (rRNA or snRNA), they then form a complex with about 4 other proteins to form RNA-protein complexes called snoRNPs. This snoRNP then catalyzes the specific modification, either 2′-O-methylation or pseudouridylation modifications. However, new research is uncovering its roles in regulating alternative splicing, regulating the levels or mRNA, and N4-acetylcytidine (ac4C) modification. 3 These regulatory functions affect cell development, which is reflected in the fact that as organismal complexity increases, so does the number of snoRNA genes. One study estimated that in the yeast Saccharomyces cerevisiae, there are 76 snoRNAs encoded in 64 transcription units, whereas in humans, the number of snoRNAs is estimated at around 550.4 Small Cajal body-specific RNAs (scaRNAs) are a new subgroup of snoRNA that only localize in the Cajal body.
snoRNAs have been implicated in a variety of cancers. Recent research shows that a specific snoRNA, SNORD1C, was highly expressed in colorectal cancer (CRC). Furthermore, knocking down SNORD1C inhibited tumor growth in vivo. The expression of SNORD1C in serum of CRC patients was significantly higher than in controls, thus positioning it as a possible biomarker from liquid biopsy samples. Similar results for a variety of different snoRNAs have revealed links between snoRNAs and lung cancer, leukemia, breast cancer, ovarian cancer, and Hepatocellular carcinoma.5
Small Nuclear RNA (snRNA)
Another class of RNAs involved in the housekeeping function of translation are snRNAs, with their main role being in splicing. These nucleosomal RNAs are localized in the eukaryotic nuclei with the splicing speckles and Cajal bodies. snRNA works in conjunction with proteins forming an RNA-protein complex called snRNPs (pronounced "snurps"), or small nuclear ribonucleoproteins. The most common human snRNA components are known U1 spliceosomal RNA, U2 spliceosomal RNA, U4 spliceosomal RNA, U5 spliceosomal RNA, and U6 spliceosomal RNA. Their nomenclature derives from their high uridine content. Within the snRNP complex, the snRNAs provide specificity to the splicing process by recognizing the sequences of critical splicing signals at the 5' and 3' ends and branch site of introns. Thus, as in rRNA, the snRNA in snRNPs perform both an enzymatic and a structural role. snRNAs are highly modified with a variety of modifications, many of them performed by snoRNAs.6
Pre-mRNA splicing is a process by which introns are removed and exons are stitched together to create the mature mRNA. The fidelity of this process is of utmost importance as exemplified by the fact that 10% of all disease-causing single-point mutations in humans generate splicing defects. snRNPs have been implicated in diseases such as Spinal muscular atrophy, Medulloblastoma, Dyskeratosis congenita, and Prader–Willi syndrome. The syndrome has been linked to the deletion of a region of paternal chromosome 15.This region includes a brain-specific snRNA that targets the serotonin-2C receptor mRNA.7
On the other side scientists are harnessing the sequence-specific splicing of snRNAs for use as therapeutic agents. For example, Locana Bio is using AAV gene therapy vectors to provide genetically engineered snRNAs with binding sites that are relevant to disease alleles. They can be used to splice out detrimental exons, or to restore protein production in frameshift mutations. 8
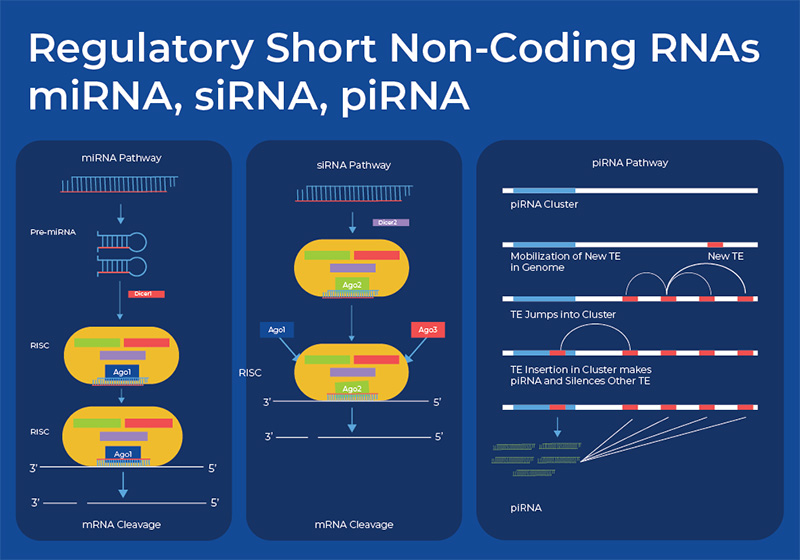
Regulatory Short non-coding RNAs
Although the snRNAs and snoRNAs are involved in the process of translation through RNA processing, modifying and splicing, we can see that they can also have regulatory roles. We will now discuss ncRNAs that are more directly involved in differential regulation of gene expression.
MicroRNA (miRNA)
MicroRNAs were first recognized as small non-coding RNAs with a regulatory function in a forward genetic screen of C. elegans in 1993. miRNAs are small non-coding RNAs, with an average length of 22 nucleotides. miRNA are exported from the nucleus as a hairpin structure that folds back upon itself. This pri-miRNA is trimmed by the Dicer enzyme, and after the other strand is discarded, the miRNA recruits the Argonaute protein and forms a RNA-Induced Silencing Complex (RISC). The single stranded miRNA is responsible for the specificity and silences the target mRNA according to base pair complementarity. In plants, this miRNA-mRNA pairing is usually completely complementary. However, in animal cells this is usually partially complementary.
In most cases, miRNA functions to suppress mRNA expression through binding to the 3’ Untranscribed region (UTR) of their target mRNA (sequence-specific manner), resulting in translational inhibition and to a lesser extent, mRNA instability. Because complete complementarity between the miRNA and its target mRNA is not required, an miRNA can target multiple mRNAs, often regulating multiple genes within a common pathway. Presently, it is believed that more than one third of the mRNA in the mammalian genome is regulated by at least one miRNA, and some mRNAs are regulated by multiple miRNAs.
miRNAs are critical components of many normal developmental pathways, and have been shown to be implicated in the development of most cells and organs. miRNAs are interwoven into transcriptional and signaling networks. However, anomalous expression has also been implicated in many diseases. Chronic lymphocytic leukemia was the first human disease shown to be caused by deregulation of miRNA.9 Since then, many other miRNAs have been associated with cancers, thus coining the term oncomirs. Expression levels of some miRNAs are shown to be upregulated in certain cancers, and can now be used as diagnostic or prognostic biomarkers. For example, in classical Hodgkin lymphoma, plasma miR-21, miR-494, and miR-1973 are promising disease response biomarkers.10 Research on miRNAs is not just revealing biomarkers of disease but also suggesting new treatment strategies. A new strategy for tumor treatment is to inhibit tumor cell proliferation by repairing the defective miRNA pathway in tumors.
Small Interfering RNA (siRNA)
Small Interfering RNAs, also known as short interfering or silencing RNA (siRNA), are double-stranded RNA molecules about 20–24 (normally 21) base pairs in length. They are similar in function to miRNA in that they silence or suppress the expression of mRNA. These RNAs are also produced by Dicer processing other RNA. After Dicer produces the siRNA duplex, other proteins are recruited and the RNA-Induced Silencing Complex (RISC) is formed. The Argonaute proteins (Ago) are a crucial component of the RISC and are directly involved in mRNA degradation. One functional difference is that siRNAs are highly-specific and have one target mRNA, whereas miRNA can regulate multiple mRNAs. Moreover, siRNAs silence gene expression by degrading mRNA, thus preventing translation. This gene-specific mechanism of knocking down genes by preventing their translation has created a variety of applied and basic research. RNA interference (RNAi) has been adopted as a functional genomics tool, and is now showing promise as a gene therapy tool. Although research is still being done on the specific delivery mechanisms and ways to minimize off target effects, four different RNAi-based therapeutics have been approved by the FDA. Each uses synthetically delivered siRNA to degrade mRNA related to a rare genetic disease11. RNAi differs from CRISPR in that RNAi blocks gene expression at the mRNA level, whereas CRISPR blocks gene expression at the DNA level.
Piwi-interacting RNA (piRNA)
Piwi-interacting RNA (piRNA) are yet another class of small non-coding RNA. piRNA is the largest class of small non-coding RNA molecules expressed in animal cells, and they are slightly longer than miRNA and siRNA at around 26-31 nt. They are similar to miRNA and siRNA in that they form a RNA-induced silencing complex (RISC) and interact with Argonaute proteins. However, rather than silencing mRNA, they are mainly involved in silencing transposable elements, although some research is revealing that they can regulate DNA methylation. piRNAs are not dependent on Dicer for biogenesis but are rather formed from transposon traps, which provide a kind of RNA-mediated adaptive immunity against transposon expansions and invasions. They also do not have any secondary structure and their function does not rely on sequence specificity. In mammals, it appears that the activity of piRNAs in transposon silencing is most important during the development of the embryo, and in both C. elegans and humans, piRNAs are necessary for spermatogenesis. 12
tRNA-derived small RNAs (tsRNA)
The newest entrant into the world of small RNAs is tRNA-derived small RNAs (tsRNA). These ncRNAs are produced by the cleavage of mature tRNAs. They act similarly to miRNAs as they form a RISC and can silence transcription through a sequence dependent manner. Moreover, they indirectly affect gene expression by acting as a sponge and binding proteins. They also regulate translation as tsRNA containing protein complexes can displace scaffolding protein eIF4G involved in translation. Also, as with all the other ncRNAs, dysregulation and modification of these tsRNAs is implicated in many diseases and cancers. They may play a large role in autoimmune disorders. One study found that 355 tsRNAs are differentially regulated in Systemic lupus erythematosus (SLE). 13
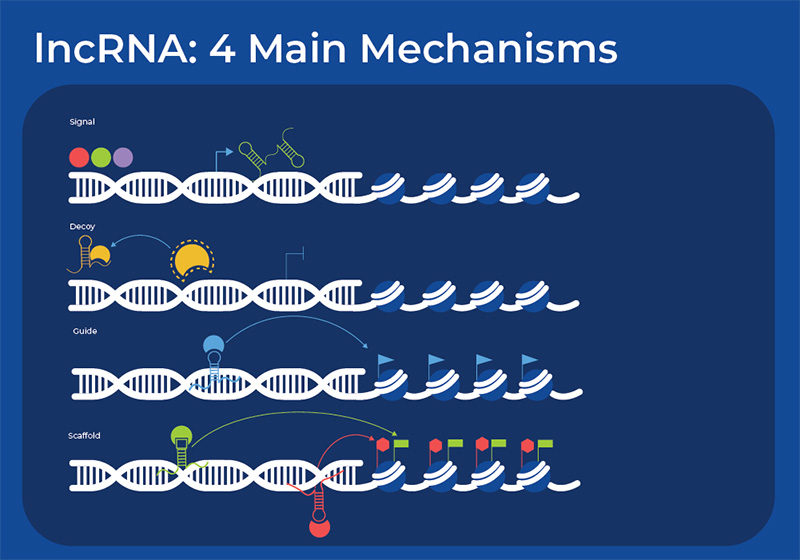
Long Non-Coding RNA (lncRNA)
Long non-coding RNA (lncRNA) encompasses the largest fraction of non-coding RNA in the genomes of complex organisms. Although a few specific cases of lncRNA had been discovered before 2000, their prevalence was unknown until large-scale transcriptomic studies indicated that the majority of the human genome was transcribed into lncRNA. Different research methods estimate that the human transcriptome contains at least 100,000 lncRNAs. Moreover, the number of lncRNAs increase with organismal complexity, which is in contrast to the fact that most eukaryotic organisms have a similar number of protein coding genes (eg. C. elegans and humans both have about 20,000 protein coding genes). Early on, many still believed that most of the lncRNAs were just transcription by-products. Since then, there has been an explosion of research revealing dynamic expression, and many biological functions for many lncRNAs. 14
The term lncRNA covers a wide diversity of RNA molecules with different biogenesis pathways, subcellular localizations, and modes of action. lncRNAs can be derived from pseudogenes, or they can be intergenic, antisense, or intronic. Similar to mRNA, most lncRNAs are transcribed by RNA polymerase II and are also polyadenylated. Additionally, loci that produce lncRNAs have many features similar to protein-coding genes, such as promoters, multiple exons, alternative splicing, chromatin signatures, regulation by morphogens and conventional transcription factors, as well as altered expression patterns observed in cancer and various other diseases. Adding to their structural diversity, lncRNAs also include circular RNAs (circRNAs) generated by back-splicing of coding and non-coding transcripts.
lncRNAs have a diversity of functions, mainly due to the fact that they have multiple structured domains that can interact with nucleic acids and proteins. There are four main mechanisms through which they function - as signals, as a decoy for other molecules, as a guide that can direct the localization of ribonucleoprotein complexes to specific targets, or as scaffolds for larger regulatory complexes. Through these mechanisms, the lncRNAs can affect biological functions at many levels: genomic stability, epigenetic, transcription, post-transcriptional regulation, and translational. In the nucleus, lncRNAs collaborate in chromatin remodeling, chromatin looping and modification, as well as control transcription and splicing. They also act as molecular sponges or decoys by binding other RNAs or proteins. One term that has been coined to describe this action is competitive endogenous RNA (ceRNA), as the lncRNA can compete with miRNA thereby de-repressing the target mRNA.
Another subclass of lncRNA is enhancer RNA (eRNA). eRNA is transcribed from the DNA sequence of enhancer regions. Recent studies suggest that eRNAs play an active role in both cis and trans transcriptional regulation, often interacting with many proteins. One well-studied eRNA is the eRNA from the enhancer that interacts with the promoter of the prostate specific antigen (PSA) gene. The PSA eRNA is strongly up-regulated by the androgen receptor and interacts directly with transcription machinery by binding to and activating the positive transcription elongation factor P-TEFb protein complex. This has a domino effect and thereby increases expression of ~ 590 androgen receptor-responsive genes. The findings indicated that PSA eRNA may be a novel target that when suppressed improves prostate cancer therapy and diagnosis. 15
lncRNAs are usually expressed in distinct cell types with expression patterns unique to each stage of differentiation and development. Approximately 80% of lncRNAs exhibit tissue specific expression, compared to 20% of mRNA. This is consistent with findings that they are important in determining cell fate and developmental trajectory. The brain contains the most diverse collection of lncRNAs with about 40% of all annotated tissue-specific lncRNAs existing in distinct brain regions. These lncRNAs function at every stage during central nervous system development and homeostasis. Advancements in this field have revealed that specific lncRNAs contribute to various brain disorders, such as Alzheimer’s disease, Parkinson’s disease, cancer, and neurodevelopmental disorders. These discoveries have spurred research around potential therapeutic interventions aimed at targeting these RNAs to restore the typical phenotype.16
The Epitranscriptome and ncRNA Interaction Networks
A new world of epitranscriptomics is being revealed in which a multitude of post-transcriptional modifications can have an impact on gene expression through affecting RNA localization, folding, stability, and decoding efficiency. Interestingly, much of this post-transcriptional modification is performed by RNAs, or RNA-protein complexes, on RNA. These RNAs that perform the modifications can be viewed as either writers, erasers, or readers of modifications. In addition to the classical role of snoRNAs and snRNAs epitranscriptomic modifications, many other ncRNAs are directly or indirectly involved in the epitranscriptome. Genetic differences in these modifying RNAs as well as environmental perturbations of them have been shown to be implicated in disease. Moreover, the epitranscriptome in the parental generation can affect organismal phenotypes in the next generation.
In addition to epitranscriptome modifications, RNA regulatory networks can become more complex due to interactions between different types of RNAs. This added layer of ncRNA-ncRNA regulation impacts epigenetic modifications, transcription, and translation. For example, experiments revealed that a circRNA, hsa_circ_0001368, decelerates the growth of gastric cancer by regulating miR-6506-5p through acting as a sponge for the miRNA. In addition to interactions between different ncRNAs, there are interactions between the same kind of ncRNAs, such as miRNA-miRNA and lncRNA-lncRNA. It has been shown that 3 distinct miRNAs - miR-125a, miR-125b, and miR-205 - act together to suppress erbB2/erbB3 expression in breast cancer cells.
Get the Complete RNA Story
Whatever your research question, if you are studying plant development, cancer, host-microbiome interactions, or developing gene therapy, it is now obvious that researchers must pay attention to the full range of transcribed RNA. These different RNA species have a variety of functions, shapes, sizes, and GC content. There is an intricate network of ncRNA’s interacting with other ncRNAs, nucleic acids, and proteins to regulate many genes and phenotypes. They are thus involved in development and disease. Norgen Biotek has a variety of RNA extraction kits that isolate total RNA, or just small RNA, with little bias toward GC content or size, thereby revealing the true RNA content of your sample. In addition to the multi-sample kit, Norgen offers specialized kits such as those for Plant/Fungi RNA, Plant miRNA, or Leukocyte RNA. Our Cytoplasmic and Nuclear RNA purification kit can also help reveal RNA localization, and there is also a single-cell RNA purification kit. Furthermore, Norgen Biotek offers a variety of sample collection and preservation methods that are designed to stabilize the true diversity of RNA in your sample.