The world as we all know it changed dramatically in the early months of 2020 with the onset of COVID-19. With business closures and hospital overloads occurring around the globe, researchers felt the urge to find solutions to the evolving pandemic. However, testing on an individual basis was soon found to be expensive, time consuming, and full of obstacles. With a lack of supplies (i.e. swabs, masks, etc.) and a shortage of healthy and cooperative health care professionals, it was quickly understood that testing every individual was not an effective or realistic solution. Luckily, researchers were able to turn to history to find viable and innovative solutions.
History of Virology
The ongoing COVID-19 pandemic, driven by SARS-CoV-2, has led to numerous innovations to aid with mass population screening and epidemiological analyses. Fortunately, researchers have been able to rely on the history of virology to help find mass screening solutions for COVID-19. In the 1990s, wastewater surveillance was introduced for mass screening as an effort to eradicate Poliovirus (Larsen & Wigginton, 2020). Poliovirus, commonly referred to as polio, was found to spread mainly through fecal-oral transmission and was linked to the onset of acute flaccid paralysis in one out of every 200 infected individuals (Larsen & Wigginton, 2020).
The introduction of wastewater surveillance was key in monitoring poliovirus outbreaks since poliovirus RNA could be detected in fecal sewage (Larsen & Wigginton, 2020). Although COVID-19 mainly spreads through respiratory droplets such as saliva, SARS-CoV-2 RNA has been detected in the stool of COVID-19 patients (Larsen & Wigginton, 2020; Peccia et al., 2020).
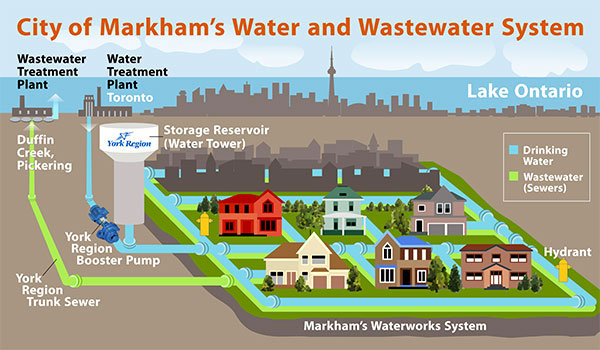 (Graphic credits to City of Markham Waterworks Dept.)
(Graphic credits to City of Markham Waterworks Dept.)
Want to hear more from Norgen?
Join over 10,000 scientists, bioinformaticians, and researchers who receive our exclusive deals, industry updates, and more, directly to their inbox.
For a limited time, subscribe and SAVE 10% on your next purchase!
SIGN UP
Current COVID-19 Testing Methods
The status quo for case reporting methods currently relies heavily on the results from tests of symptomatic, ill individuals. The current testing procedure involves nasopharyngeal swab or saliva sample collection, viral RNA isolation, RT-qPCR for viral detection, and reporting. Testing is administered to individuals who have self-reported symptoms or who have been diagnosed by a physician. Unfortunately, due to long wait times and a myriad of individuals requiring testing, the current reporting method for COVID-19 has not been 100% effective. As a result, other environmental monitoring methods have been suggested to help understand the development of ongoing and future pandemics.
Wastewater Surveillance
Sampling and testing of wastewater has been suggested as a method to track the disease dynamics of COVID-19 (Peccia et al., 2020; Wu et al., Preprint; Ahmed et al., 2020). SARS-CoV-2 RNA has been detected in a variety of wastewater types (primary sludge, composite samples and other untreated wastewater) (Ahmed et al., 2020; Peccia et al., 2020). In 2020, Peccia et al. tested primary sewage sludge for SARS-CoV-2 and were able to detect SARS-CoV-2 over a 10-week period. They identified that their detection of SARS-CoV-2 was 6 to 8 days ahead of the reporting date for clinical tests. The trends in their detection were similar to the clinical case trends of increases and decreases that occurred during the 10-week period, although their trends were detectable sooner than the clinical cases.
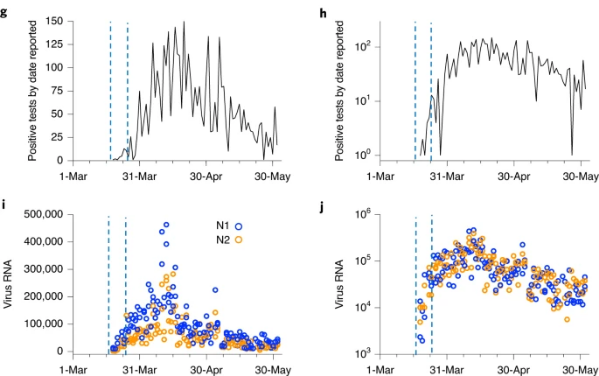 Figure 1. Sludge SARS-CoV-2 RNA concentration time course and COVID-19 outbreak indicators on linear (left) and log (right) scales. g,h, Number of positive SARS-CoV-2 test results by public reporting date, i,j, Primary sludge SARS-CoV-2 RNA concentration (virus RNA gene copies per ml of sludge) (Peccia et al., 2020).
Figure 1. Sludge SARS-CoV-2 RNA concentration time course and COVID-19 outbreak indicators on linear (left) and log (right) scales. g,h, Number of positive SARS-CoV-2 test results by public reporting date, i,j, Primary sludge SARS-CoV-2 RNA concentration (virus RNA gene copies per ml of sludge) (Peccia et al., 2020).
These results are echoed by other findings from Wu et al. (Preprint) and Ahmed et al. (2020), recommending wastewater surveillance to track SARS-CoV-2 and other infectious diseases that are excreted in feces. Wastewater surveillance provides researchers with another epidemiological tool that can help advise on health measures, 6 to 8 days prior to publicly reported data.
Norgen’s Solution
 We at Norgen have developed a supplementary protocol for our Soil Total RNA Purification Kit in response to the increasing need for environmental monitoring and mass screening. This supplementary protocol can be used for the purification of viral RNA from complex wastewater samples, using a Centricon® Plus-70 Centrifugal Filter and then isolation with our Soil Total RNA Purification Kit (product #27750).
We at Norgen have developed a supplementary protocol for our Soil Total RNA Purification Kit in response to the increasing need for environmental monitoring and mass screening. This supplementary protocol can be used for the purification of viral RNA from complex wastewater samples, using a Centricon® Plus-70 Centrifugal Filter and then isolation with our Soil Total RNA Purification Kit (product #27750).
If you have any recommendations for a blog topic, comment your ideas below!