According to the World Health Organization, cancer was the leading cause of death worldwide in 2020 1. With countless individuals dying from breast, lung, colorectal and other forms of cancers daily, researchers have dug deep into the world of circulating tumour DNA (ct-DNA) to learn more about early disease diagnosis and treatment. Although cancer remains mostly incurable, it can be treated, and when caught early, chances of survival dramatically increase. In this blog, we'll cover what ct-DNA is and how it could be a game-changer for cancer diagnosis, surveillance, and treatment.
The ct-DNA Advantage
 Over the past century, tumour detection and early cancer diagnosis have been done through imaging of a body part or by taking a tissue biopsy and examining it for histological signatures of tumours 2. These limited and invasive methods rely on the presence of clinically detectable tumours and are surgically accessible for biopsy collection. Fortunately, researchers can now overcome these limitations due to the discovery of ct-DNA in bodily fluids. ct-DNA, which is a type of cell free-DNA, is shed by tumours through apoptosis, necrosis, or active transport in vesicles2.
Over the past century, tumour detection and early cancer diagnosis have been done through imaging of a body part or by taking a tissue biopsy and examining it for histological signatures of tumours 2. These limited and invasive methods rely on the presence of clinically detectable tumours and are surgically accessible for biopsy collection. Fortunately, researchers can now overcome these limitations due to the discovery of ct-DNA in bodily fluids. ct-DNA, which is a type of cell free-DNA, is shed by tumours through apoptosis, necrosis, or active transport in vesicles2.
NORBLOG
Want to hear more from Norgen?
Join over 10,000 scientists, bioinformaticians, and researchers who receive our exclusive deals, industry updates, and more, directly to their inbox.
For a limited time, subscribe and SAVE 10% on your next purchase!
SIGN UP
The use of ct-DNA as a primary method of tumour detection and diagnosis offers a variety of advantages:
-
Early detection of tumours that are as small as 11 mm in diameter (3).
-
Opportunity for early cancer detection and therefore earlier treatment.
-
Cancer detection sensitivity from 50-70% (3).
-
Relies on noninvasive, easily harvested, repeatable biopsies of bodily fluids (2).
-
Allows for the determination of the temporal genetic change and heterogeneity of the tumour (4).
Cancer Diagnosis and Therapy
 To better understand these advantages, we will take the fictional example of Dr. Sakari and her patients, Sophia and Étienne.
The advantages of ct-DNA led Dr. Sakari from Iqaluit, Nunavut, to use liquid biopsies for two patients that were referred to her clinic for cancer diagnosis and therapy. The first patient, Sophia, was concerned about having breast cancer. The second patient, Étienne, had recently been diagnosed with stage III pancreatic cancer. Dr. Sakari had to determine the type of liquid biopsies to use in the diagnosis and treatment of their cancers.
To better understand these advantages, we will take the fictional example of Dr. Sakari and her patients, Sophia and Étienne.
The advantages of ct-DNA led Dr. Sakari from Iqaluit, Nunavut, to use liquid biopsies for two patients that were referred to her clinic for cancer diagnosis and therapy. The first patient, Sophia, was concerned about having breast cancer. The second patient, Étienne, had recently been diagnosed with stage III pancreatic cancer. Dr. Sakari had to determine the type of liquid biopsies to use in the diagnosis and treatment of their cancers.
MAFs & the Two Ss of Cancer Diagnosis
 The type of liquid biopsy needed to diagnose and treat cancer depends on the known or suspected location of the tumour; the closer proximity of the liquid biopsy to the tumour, the higher likelihood of isolating ct-DNA from that tumour 5. Dr. Sakari asked Étienne for a urine sample because of the close proximity of the pancreas to the urethra. As for Sophia, a breast milk sample would have been preferred, but because she is not lactating, a blood sample was collected instead. After obtaining liquid biopsies from her patients, Dr. Sakari sent the samples for cf-DNA isolation, quantification, and next-generation sequencing of ct-DNA. Based on the sequencing information, Dr. Sakari estimates the mutant allele fraction (MAF) in each patient's biopsy. MAF is the fraction of mutant alleles to the wild-type alleles found in the biopsy 6 which can be used in two different ways for staging cancer:
The type of liquid biopsy needed to diagnose and treat cancer depends on the known or suspected location of the tumour; the closer proximity of the liquid biopsy to the tumour, the higher likelihood of isolating ct-DNA from that tumour 5. Dr. Sakari asked Étienne for a urine sample because of the close proximity of the pancreas to the urethra. As for Sophia, a breast milk sample would have been preferred, but because she is not lactating, a blood sample was collected instead. After obtaining liquid biopsies from her patients, Dr. Sakari sent the samples for cf-DNA isolation, quantification, and next-generation sequencing of ct-DNA. Based on the sequencing information, Dr. Sakari estimates the mutant allele fraction (MAF) in each patient's biopsy. MAF is the fraction of mutant alleles to the wild-type alleles found in the biopsy 6 which can be used in two different ways for staging cancer:
Total MAF
Since tumour formation occurs through increased uncontrolled cell proliferation, each round of cell replication introduces more mutations into the tumour, which appear in the ct-DNA. This means that a small tumour composed of only a few cells contains a small number of mutations, while a large tumour contains a large number of mutations. Therefore, the amount of total mutant alleles to total wild-type alleles should be lower in ct-DNA samples taken from a small tumour than a large one. It has been shown that MAF can be used to detect tumours that are as small as 11 mm in diameter 3.
MAF of Driver Genes
Another use of MAFs for tumour detection is coupled with the use of driver genes, which are used as cancer biomarkers. Driver genes include oncogenes as well as tumour-suppressor genes 7 and play important roles in cancer formation. The utility of a specific driver gene depends on two crucial variables, being sensitivity and specificity (the two ‘Ss' of cancer diagnosis). Sensitivity is the percentage of correctly identified patients with cancer (true positive rate), while specificity is the proportion of correctly identified patients without cancer (false positive rate) 5. The perfect biomarker would have 100% specificity and sensitivity, but unfortunately, this does not exist. A useful biomarker can correctly assign a state of presence or absence of cancer in a person's body more accurately than random assignment.
Sophia's Diagnosis
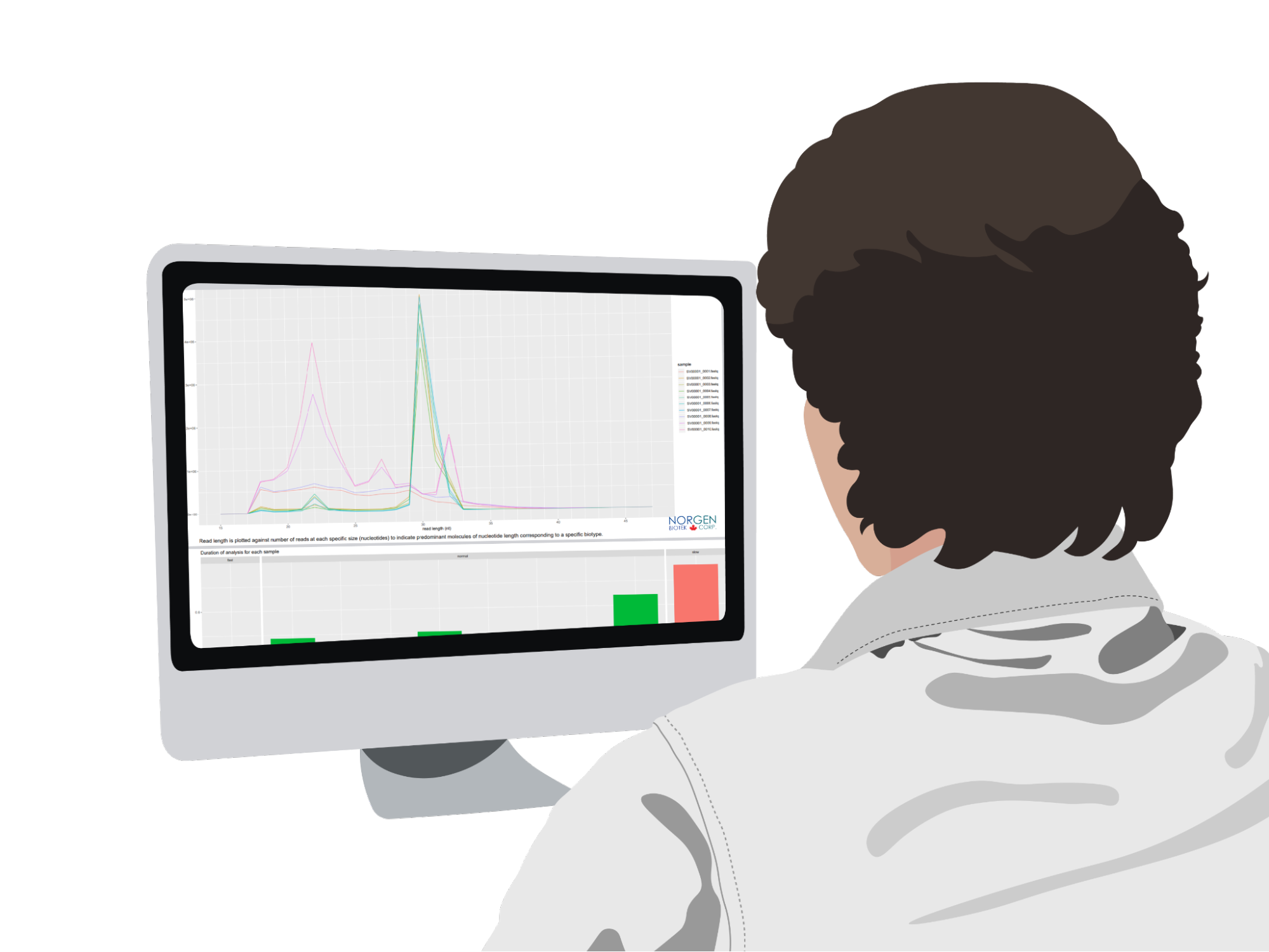 Dr. Sakari sent Sophia's blood sample to a lab to conduct a variety of assays on it. The lab technicians separated the plasma from the blood and quantified the amount of cf-DNA in it. The quantification was done using RT-PCR and found that she has 17 ng per ml in her plasma which is 10 times higher than the average 7 ng per ml in a healthy person's plasma 8. Afterwards, the lab technicians used droplet digital polymerase chain reaction (ddPCR) to quantify the mutant allele for the MAF ratio and examine any mutations in breast cancer driver genes. ddPCR is a type of PCR reaction that is able to detect and quantify mutations in DNA samples. After ddPCR, next-generation sequencing was performed to determine the exact mutations in Sophia's cf-DNA. The lab results showed that Sophia's plasma had a higher MAF than what is typically found in healthy individuals. They also found that a tumorigenic mutation in the Phosphatidylinositol-4,5-Bisphosphate 3-Kinase Catalytic Subunit Alpha (PIK3CA) gene is present which codes for PIK protein that regulates cell proliferation. PIK3CA mutations lead to the increase of cell proliferation and tumour formation 9 and have been shown to be a biomarker for breast cancer with a 93% sensitivity and 100% specificity 10.
Dr. Sakari sent Sophia's blood sample to a lab to conduct a variety of assays on it. The lab technicians separated the plasma from the blood and quantified the amount of cf-DNA in it. The quantification was done using RT-PCR and found that she has 17 ng per ml in her plasma which is 10 times higher than the average 7 ng per ml in a healthy person's plasma 8. Afterwards, the lab technicians used droplet digital polymerase chain reaction (ddPCR) to quantify the mutant allele for the MAF ratio and examine any mutations in breast cancer driver genes. ddPCR is a type of PCR reaction that is able to detect and quantify mutations in DNA samples. After ddPCR, next-generation sequencing was performed to determine the exact mutations in Sophia's cf-DNA. The lab results showed that Sophia's plasma had a higher MAF than what is typically found in healthy individuals. They also found that a tumorigenic mutation in the Phosphatidylinositol-4,5-Bisphosphate 3-Kinase Catalytic Subunit Alpha (PIK3CA) gene is present which codes for PIK protein that regulates cell proliferation. PIK3CA mutations lead to the increase of cell proliferation and tumour formation 9 and have been shown to be a biomarker for breast cancer with a 93% sensitivity and 100% specificity 10.
With this information, Dr. Sakari concluded that Sophia's suspicion about having breast cancer was correct and planned an imaging and tissue biopsy session with Sophia to confirm the presence of cancer.
Étienne's Cancer Therapy
Étienne has already been diagnosed with stage III pancreatic cancer. Dr. Sakari wanted to determine the location of the cancer mutations in Étienne's tumour and check if the tumour would be responsive to therapy. She requested a urine sample from Étienne and sent it to the lab to analyze the ct-DNA in it. The lab technicians isolated the DNA from the sample and used ddPCR to determine the number of mutated alleles of the gene Kirsten RA Sarcoma (KRAS) 11. KRAS is an oncogene that controls the activation of cellular growth factors and consequently, cell proliferation. The lab technicians found that three different mutated alleles have a high frequency in Étienne's urine sample. When the lab sent these results to Dr. Sakari, she started Étienne on a course of medication to treat his cancer. She requested new urine samples every 14 days to track the effects of Étienne's therapy. The continual sampling of urine provides a way for Dr. Sakari to determine if Étienne's tumour is regressing. Indeed, she found that the frequency of KRAS mutant alleles decreased after a year of medication, meaning that Étienne's cancer is in remission.
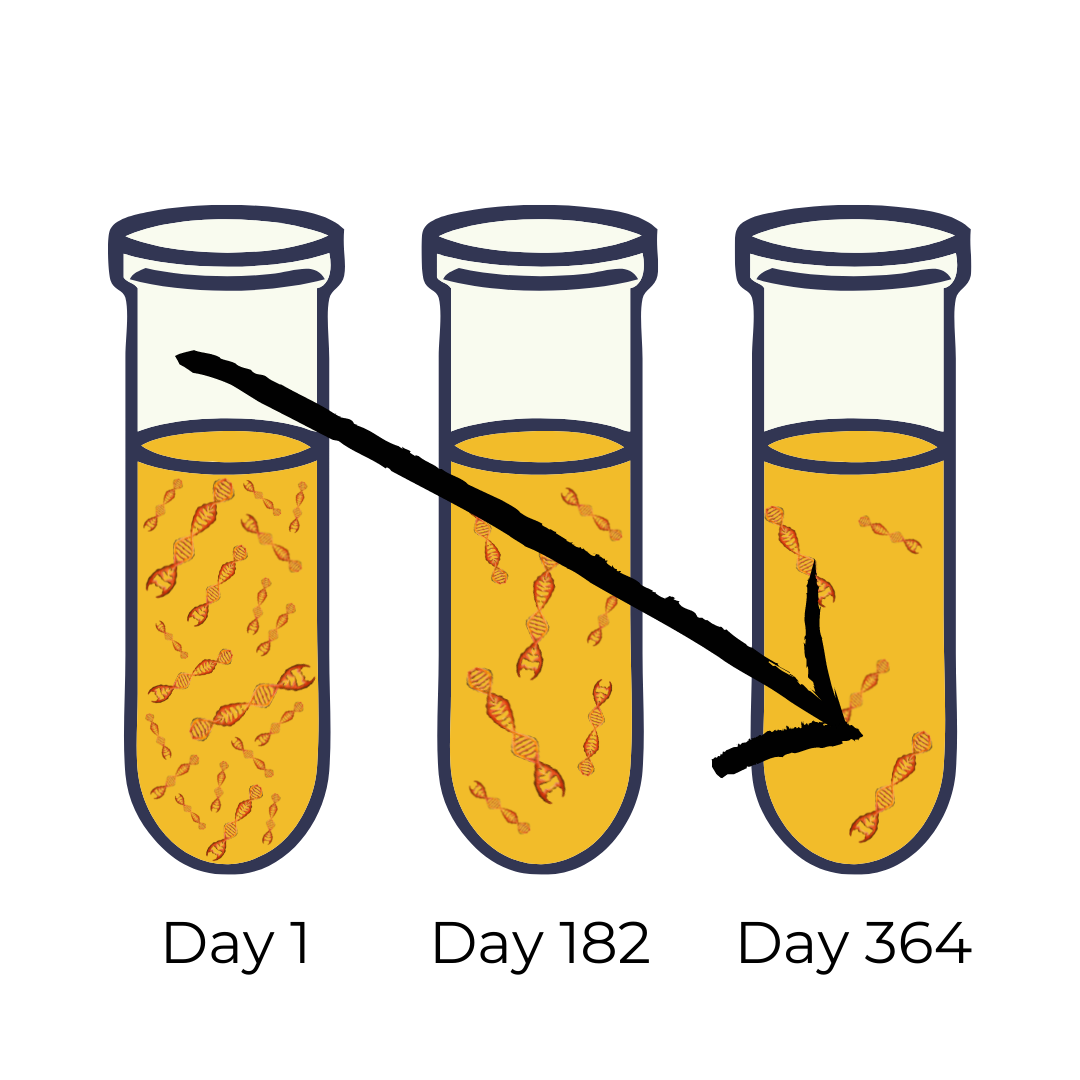 ct-DNA is a type of cf-DNA that has become an invaluable tool for cancer diagnosis and therapy. The use of ct-DNA for cancer diagnosis has a variety of advantages over traditional methods, which stem from its ease of collection and recollection and the content of the ct-DNA. ct-DNA houses mutant alleles, whose quantity and sequence can be used to diagnose cancer (as in the case of Sophia) and tracking the effectiveness of cancer treatment (as in the case of Étienne).
ct-DNA is a type of cf-DNA that has become an invaluable tool for cancer diagnosis and therapy. The use of ct-DNA for cancer diagnosis has a variety of advantages over traditional methods, which stem from its ease of collection and recollection and the content of the ct-DNA. ct-DNA houses mutant alleles, whose quantity and sequence can be used to diagnose cancer (as in the case of Sophia) and tracking the effectiveness of cancer treatment (as in the case of Étienne).